12.3 The component hierarchy
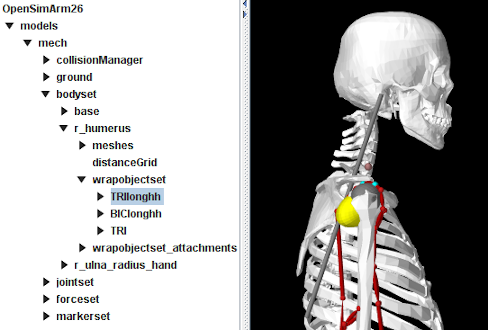
OpenSimParser arranges implementing components within MechModel in a manner that preserves the hierarchy and naming of the original OpenSim model. This results in a component hierarchy different from the canonical one used by MechModel, in which components are placed in container lists by type (e.g., particles in a PointList<Particle> named "particles", rigid bodies in a RenderableComponentList<RigidBody> named "rigidBodies", axial springs in an AxialSpringList<AxialSpring> list named "axialSprings"). However, as described in Section 4.8, applications are free to create custom component arrangements, and OpenSimParser does this, recreating the basic OpenSim containers "bodyset", "jointset" (for OpenSim 4), "forceset", "constraintset" and "markerset". Figure 12.2 illustrates this by showing the ArtiSynth navigation panel for the OpenSimArm26 example. The only original MechModel component that appears is the collisionManager (the other canonical containers, such as "particles", "rigidBodies", etc., are still present, but don’t appear because they are empty).
12.3.1 Ground
If present, the OpenSim 4 "ground" component appears as its own component, implemented using a RigidBody whose dynamic property is set false and grounded property is set true.
12.3.2 Body set
The body set is implemented as a RenderableComponentList<RigidBody>, with each body implemented by a special implementation of RigidBody that contains a child component named "wrapobjectset" that in turn contains any attached wrap objects. Wrap objects themselves are implemented by special instances of RigidBody that implement the WrapComponent interface. Figure 12.2 shows the navigation panel with the body r_humerus expanded and the wrap object TRIlonghh selected. Other body components include meshes and distancegrid, which are defined for all ArtiSynth rigid bodies, and wrapobjectset_attachments, which are the internal attachments connecting the wrap objects to the body.
For OpenSim 3 models, each body also contains a child component "joint", containing the single joint for which that body is the parent. This is in place of the joint set provided by OpenSim 4.
12.3.3 Joint set
For OpenSim 4, the joint set is implemented as a RenderableComponentList<JointBase> containing JointBase components realizing the various OpenSim joints. Custom joints, in particular, are implemented by OpenSimCustomJoint.
12.3.4 Force set
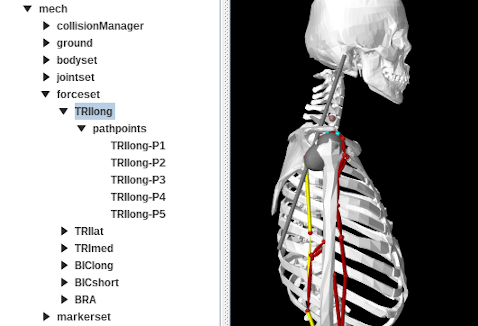
The force set is implemented as a RenderableComponentList<ModelComponent> containing all the supported force effectors. Bushings and CoordinateLimitForces are realized using FrameSpring and JointLimitForce. CoordinateActuators are implemented using JointActuator, while PointToPointActuator is implemented using a custom component TwoPointActuator. Muscles and point-to-point springs are implemented using a special implementation of MultiPointMuscle that contains a child component named "pathpoints" that in turn contains all the muscle’s path points. Each path point is usually a FrameMarker attached to one of the bodies in the body set. Figure 12.3 shows the navigation panel with the muscle TRIlong selected and also expanded to show the path points.
Currently supported OpenSim muscles include Thelen2003Muscle and Millard2012EquilibriumMuscle, which are implemented using the MultiPointMuscle materials Thelen2003AxialMuscle and Millard2012AxialMuscle (Section 4.5.4). All other OpenSim muscles are implemented as a generic instance of ConstantAxialMuscle (Section 4.5.1) whose maxForce property is set to the specified max_isometric_force.
12.3.5 Constraint set
The constraint set is implemented as a ComponentList<ConstrainerBase> containing all the supported constrainers. At present, these include CoordinateCouplerConstraint and PointConstraint, realized by JointCoordinateCoupling and SphericalJoint, respectively.
12.3.6 Marker set
The marker set is implemented as a PointList<Marker> with each marker realized using FrameMarker.